Red Risk Level
HIGH RISK
Fresh out of the COVID pandemic we’ve come to dread our mutations. We test for signs of mutation with fear and loathing. It’s past time we corrected our attitude toward this inevitable and constant process.
Mutation drives evolution. Evolution proceeds through mutation.
The force that steered us from a future as a puddle of amino acids or a bacteria flagelatting around without a nucleus or purpose is mutation.
Everything changes constantly. And the engine of change in biological systems is: Mutation.
This little almanac is a description of a handful of the most common driving and passenger MPN mutations currently discovered. These are those genetic changes that create the proteins that play a role in the swelling, itching, inflammation and fatigue that characterize the myeloproflierative chaotic blood production system. (Hematopoiesis.)
The Almanac is designed to be used with the Myelofibrosis Risk Assessment Tool (www.mrat.com) although it can service as an introduction for all the MPN phenotypes.
There are many more mutations than those treated here, some likely more significant, but these, lab tested and verified, may do for a start.
Mutations arise in the course of cellular reproduction, a complex and decidedly unsexy process of spindles and splitting, chromosomes and transcription to messenger RNA and translation to proteins via ribosomes cruising our cytoplasm before getting folded into a workable, functional proteins. Trillions of cells continually reproducing their yards long alphabet soup of DNA with the inevitable errors – changes — arising from copying, moving, shifting, etc.
Mutations occur within genes, clearly defined relatively short sections of DNA. Sometime they take off expand, usually not. And unless the mutation occurs within the germline – egg and sperm cells –they are somatic and not passed along to our kids. Our genome is made up of two long strips of DNA, one from Mom, one from Dad. If a mutation occurs in a gene on one strip only it’s homozygous. If it occurs on both strips its’ heterozygous.
Which gets us to allele burden. The percentage of a mutated gene in our genome is the allele burden. So if you’re homozygous for a mutation your maximum allele burden is 50%. Heterozygous? 100%. Unmutated genes are called WT or Wild Type.
Good news: Some mutations are good for us, help assure our survival. Most mutations, which occur in single cells don’t last very long or are otherwise of little significance. But the bad news is a few are downright nasty and if they include a survival advantage, they can increase in number and power and create problems.
Finally, in our Little Almanac of MPN Mutations each mutation is listed alphabetically, again to make it easier to find and discuss with your hematologist. We start each description with the location of the mutated gene along its chromosome. The purpose is to help familiarize us with the mutation and thus impress our physicians when we discuss its presence but mostly to illustrate how long a gene may be and how the impact of a particular mutation depends on where along the sprink;ing of Letters (bases) the mutation has occurred. All ASLX-1 mutations are not equal.
The bases – nucleobases –Adenine (A), Guanine (G) Thymine (T) Cytosine (C) and Uracil (U)) compounds are the nitrogen containing molecules that make up the twisted double helix DNA or RNA ladder (G) is always paired with (C) and (A) is paired with (U) or (T).

The most common MPN mutations
Rank and Type

The Mutations….
This is an alphabetic listing of the 15 most prevalent genes based on the Grinfeld,Nangalia study;.survey of MPN patients across all MPN phenotypes.N=2005,)
ASLX1 (Ranking: 3, 21%)
(ASLX Transcriptional regulator)
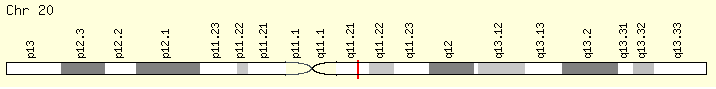
The ASXL1 gene provides instructions for making a protein that is involved in a process known as chromatin remodeling. Chromatin is the complex of DNA and proteins that packages DNA into chromosomes. The ASXL1 protein may have an additional role in gene regulation by signaling to molecules to add a methyl group (a process called methylation) to an area near a gene called the promoter region, which controls gene activity. When a promoter region is methylated, gene activity is repressed, and when a promoter region is not methylated, the gene is active. https://ghr.nlm.nih.gov/gene/ASXL1
ASLX1 mutation correction by CRISPR/Cas9 restores gene function in leukemia cells. (Valletta, S. et al. Oncotarget, 2015)
CALR (Ranking: 2, 32%)
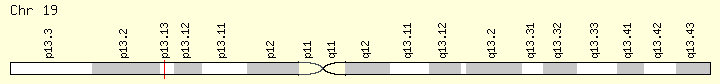
The CALR gene provides instructions for making a multi-functional protein called calreticulin. This protein is found in several parts of the cell, including inside a structure called the endoplasmic reticulum (ER), in the fluid-filled space inside the cell (the cytoplasm), and at the outer surface of the cell. The ER is involved in protein processing and transport, and within this structure, calreticulin plays a role in ensuring the proper folding of newly formed proteins.
The CALR gene provides instructions for making a multi-functional protein called calreticulin. This protein is found in several parts of the cell, including inside a structure called the endoplasmic reticulum (ER), in the fluid-filled space inside the cell (the cytoplasm), and at the outer surface of the cell. The ER is involved in protein processing and transport, and within this structure, calreticulin plays a role in ensuring the proper folding of newly formed proteins. The ER is also a storage location for charged calcium atoms (calcium ions), and calreticulin is involved in maintaining the correct levels of calcium ions in this structure. Through calcium regulation and other mechanisms, calreticulin is thought to play a role in the control of gene activity, cell growth and division (proliferation) and movement (migration), the attachment of cells to one another (adhesion), and regulation of programmed cell death (apoptosis). The function of this protein is important for immune system function and wound healing. https://ghr.nlm.nih.gov/gene/CALR
CBL (Ranking: 10, 6%)
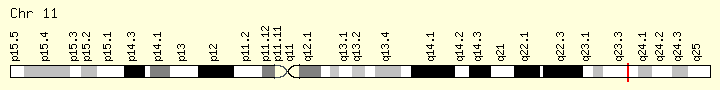
Cbl (named after Casitas B-lineage Lymphoma) is a mammalian gene encoding the protein CBL which is an E3 ubiquitin-protein ligase involved in cell signalling and protein ubiquitination. Mutations to this gene have been implicated in a number of human cancers, particularly acute myeloid leukaemia
Ubiquitination is the process of chemically attaching ubiquitin monomers to a protein, thereby targeting it for degradation. As this is a multi-step process, several different enzymes are involved, the final one being a member of the E3 family of ligases. Cbl functions as an E3 ligase, and therefore is able to catalyse the formation of a covalent bond between ubiquitin and Cbl’s protein substrate – typically a receptor tyrosine kinase.
This gene is a proto-oncogene that encodes a RING finger E3 ubiquitin ligase. The encoded protein is one of the enzymes required for targeting substrates for degradation by the proteasome. This protein mediates the transfer of ubiquitin from ubiquitin conjugating enzymes (E2) to specific substrates. This protein also contains an N-terminal phosphotyrosine binding domain that allows it to interact with numerous tyrosine-phosphorylated substrates and target them for proteasome degradation. As such it functions as a negative regulator of many signal transduction pathways. This gene has been found to be mutated or translocated in many cancers including acute myeloid leukaemia, and expansion of CGG repeats in the 5′ UTR has been associated with Jacobsen syndrome. Mutations in this gene are also the cause of Noonan syndrome-like disorder. [provided by RefSeq, Jul 2016] https://ghr.nlm.nih.gov/gene/CBL
DNMT3A (Ranking: 8, 7%)
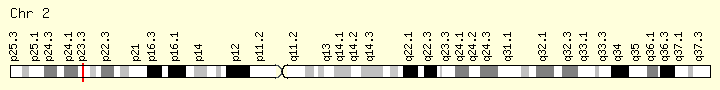
DNA Methyl Transferase 3 Alpha
The DNMT3A gene provides instructions for making an enzyme called DNA methyltransferase 3 alpha. This enzyme is involved in DNA methylation, which is the addition of methyl groups, consisting of one carbon atom and three hydrogen atoms, to DNA molecules. In particular, the enzyme helps add methyl groups to DNA building blocks (nucleotides) called cytosines.
DNA methylation is important in many cellular functions. These include determining whether the instructions in a particular segment of DNA are carried out or suppressed (gene silencing), regulating reactions involving proteins and fats, and controlling the processing of chemicals that relay signals in the nervous system (neurotransmitters). DNA methyltransferase 3 alpha is particularly important for establishing DNA methylation patterns during development before birth. The enzyme also functions in early cells that can give rise to more mature cell types. In early blood cells, called hematopoietic stem cells, the methylation patterns established by DNA methyltransferase 3 alpha promote maturation (differentiation) into different blood cell types.
Mutations in the DNMT3A gene are associated with a form of blood cancer known as cytogenetically normal acute myeloid leukemia (CN-AML). While large chromosomal abnormalities can be involved in the development of acute myeloid leukemia, about half the cases do not have these abnormalities; these are classified as CN-AML. Up to one-third of people with CN-AML have a mutation in the DNMT3A gene.
DNA methylation plays an important role in animal development and gene regulation. In mammals, several genes encoding DNA cytosine methyltransferases have been identified. DNMT1 is constitutively expressed and is required for the maintenance of global methylation after DNA replication
DNMT3A at 6.8% occurrence is the 8th most common MPN mutation https://ghr.nlm.nih.gov/gene/DNMT3A
EZH2 (Ranking: 14M 4%)
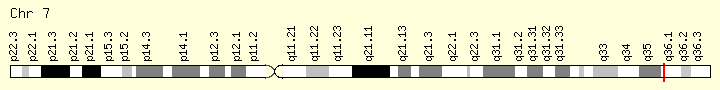
Enhancer of Zeste 2 polycomb repressive complex
The EZH2 gene provides instructions for making a type of enzyme called a histone methyltransferase. Histone methyltransferases modify proteins called histones, which are structural proteins that attach (bind) to DNA and give chromosomes their shape. By adding a molecule called a methyl group to histones (methylation), histone methyltransferases can turn off (suppress) the activity of certain genes, an essential process in normal development. Specifically, the EZH2 enzyme forms part of a protein group called the polycomb repressive complex-2. By turning off particular genes, this complex is involved in the process that determines the type of cell an immature cell will ultimately become (cell fate determination).
IDH2\DLT3 (Ranking: 11, 5%)

Isocitrate Dehydrogenase 2, Mitochondrial
The IDH2 gene provides instructions for making an enzyme called isocitrate dehydrogenase 2. This enzyme is found in mitochondria, which are the energy-producing centers within cells. Within mitochondria, the enzyme participates in reactions that produce energy for cell activities. Specifically, isocitrate dehydrogenase 2 normally converts a compound called isocitrate to another compound called 2-ketoglutarate. A series of additional enzymes further process 2-ketoglutarate to produce energy. The conversion reaction also produces a molecule called NADPH, which is necessary for many cellular processes and helps protect cells from potentially harmful molecules called reactive oxygen spe
JAK2 (Ranking: 1, 66%)cies. https://ghr.nlm.nih.gov/gene/IDH2
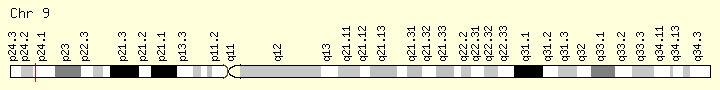
Janus Kinase 2
The JAK2 gene provides instructions for making a protein that promotes the growth and division (proliferation) of cells. This protein is part of a signaling pathway called the JAK/STAT pathway, which transmits chemical signals from outside the cell to the cell’s nucleus. The JAK2 protein is especially important for controlling the production of blood cells from hematopoietic stem cells. These stem cells are located within the bone marrow and have the potential to develop into red blood cells, white blood cells, and platelets https://ghr.nlm.nih.gov/gene/JAK2
KMT2A (Ranking: 9, 6%)
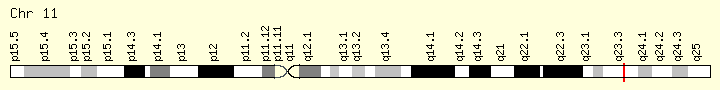
Lysine methyltransferase
This gene encodes a transcriptional coactivator that plays an essential role in regulating gene expression during early development and hematopoiesis. The encoded protein contains multiple conserved functional domains. One of these domains, the SET domain, is responsible for its histone H3 lysine 4 (H3K4) methyltransferase activity which mediates chromatin modifications associated with epigenetic transcriptional activation. This protein is processed by the enzyme Taspase 1 into two fragments, MLL-C and MLL-N. These fragments reassociate and further assemble into different multiprotein complexes that regulate the transcription of specific target genes, including many of the HOX genes. Multiple chromosomal translocations involving this gene are the cause of certain acute lymphoid leukemias and acute myeloid leukemias. Alternate splicing results in multiple transcript variants.[provided by RefSeq, Oct 2010] https://ghr.nlm.nih.gov/gene/KMT2A
MPL (Ranking: 6, 7%)
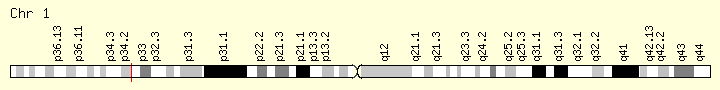
Myeloprofliferative Proto Oncogene Thrombopoietin Receptor
Like essential thrombocythemia, primary myelofibrosis is associated with the MPL gene mutations referred to as W515 mutations. These mutations lead to a constitutively activated thrombopoietin receptor protein, which results in the overproduction of abnormal megakaryocytes. These megakaryocytes stimulate other cells to release collagen, a protein that normally provides structural support for the cells in the bone marrow but causes scar tissue formation in primary myelofibrosis. Because of the fibrosis, the bone marrow cannot produce enough normal blood cells, leading to the signs and symptoms of the condition.
In 1990 an oncogene, v-mpl, was identified from the murine myeloproliferative leukemia virus that was capable of immortalizing bone marrow hematopoietic cells from different lineages. In 1992 the human homologue, named, c-mpl, was cloned. Sequence data revealed that c-mpl encoded a protein that was homologous with members of the hematopoietic receptor superfamily
SRSF2 (Ranking: 5, 8%)
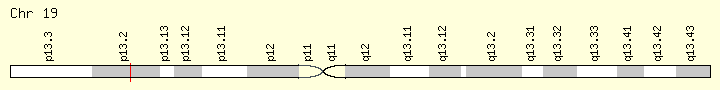
Second Step Splicing Factor 2 ( Peter Pan homolog)
The protein encoded by this gene is a member of the serine/arginine (SR)-rich family of pre-mRNA splicing factors, which constitute part of the spliceosome. Each of these factors contains an RNA recognition motif (RRM) for binding RNA and an RS domain for binding other proteins. The RS domain is rich in serine and arginine residues and facilitates interaction between different SR splicing factors. In addition to being critical for mRNA splicing, the SR proteins have also been shown to be involved in mRNA export from the nucleus and in translation. Two transcript variants encoding the same protein and one non-coding transcript variant have been found for this gene. In addition, a pseudogene of this gene has been found on chromosome 11. [provided by RefSeq, Sep 2010] https://ghr.nlm.nih.gov/gene/SRSF2
SF3B1 (Ranking: 7, 7%)
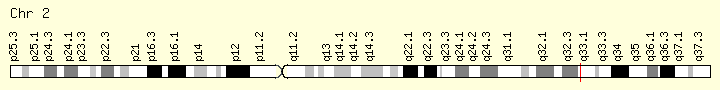
Splicing Factor 3B Subunit 1
Splicing Factor 3B1
The SF3B1 gene provides instructions for making the protein, which is part of a complex called a spliceosome. Spliceosomes help process messenger RNA (mRNA), which is a chemical cousin of DNA that serves as a genetic blueprint for making proteins. The spliceosomes recognize and then remove regions from mRNA molecules that are not used in the blueprint (which are called introns).
Mutations in this gene have been recurrently seen in cases of advanced chronic lymphocytic leukemia,[12] myelodysplastic syndromes[13] and breast cancer.[14] SF3B1 mutations are found in 60%-80% of patients with refractory anemia with ring sideroblasts (RARS; which is a myelodysplastic syndrome) or RARS with thrombocytosis (RARS-T; which is a myelodysplastic syndrome/myeloproliferative neoplasm). There is also an emerging body of evidence to suggest implications of SF3B1 mutations being involved in orbital melanoma. https://ghr.nlm.nih.gov/gene/SF3B1
TET 2 (Ranking: 4, 19%)

TET methylcytosine dioxygenase 2
The TET2 gene provides instructions for making a protein whose function is unknown. Based on the function of similar proteins, researchers believe the TET2 protein is involved in regulating the process of transcription, which is the first step in protein production. Although this protein is found throughout the body, it may play a particularly important role in the production of blood cells from hematopoietic stem cells. These stem cells are located within the bone marrow and have the potential to develop into red blood cells, white blood cells, and platelets. The TET2 protein appears to act as a tumor suppressor, which is a protein that prevents cells from growing and dividing in an uncontrolled way.
Although this protein is found throughout the body, it may play a particularly important role in the production of blood cells from hematopoietic stem cells. The TET2 protein appears to act as a tumor suppressor, which is a protein that prevents cells from growing and dividing in an uncontrolled way. https://ghr.nlm.nih.gov/gene/TET2
TP53 Genomic Sub=group #1 (Ranking:19, 2%)
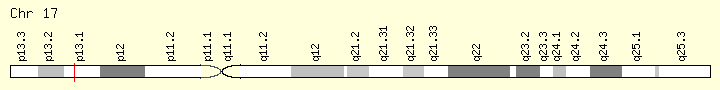
Tumor Protein P53
The TP53 gene provides instructions for making a protein called tumor protein p53 (or p53). This protein acts as a tumor suppressor, which means that it regulates cell division by keeping cells from growing and dividing (proliferating) too fast or in an uncontrolled way.
The p53 protein is located in the nucleus of cells throughout the body, where it attaches (binds) directly to DNA. When the DNA in a cell becomes damaged by agents such as toxic chemicals, radiation, or ultraviolet (UV) rays from sunlight, this protein plays a critical role in determining whether the DNA will be repaired or the damaged cell will self-destruct (undergo apoptosis). If the DNA can be repaired, p53 activates other genes to fix the damage. If the DNA cannot be repaired, this protein prevents the cell from dividing and signals it to undergo apoptosis. By stopping cells with mutated or damaged DNA from dividing, p53 helps prevent the development of tumors.
Because p53 is essential for regulating cell division and preventing tumor formation, it has been nicknamed the “guardian of the genome.”
Cytogenetic Location: 17p13.1, which is the short (p) arm of chromosome 17 at position 13.1
Molecular Location: base pairs 7,668,402 to 7,687,550 on chromosome 17 (Homo sapiens Annotation Release 109, GRCh38.p12) (NCBI)
U2AF1 (Ranking: 13, 4%)
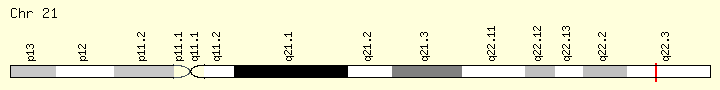
Small Nuclear RNA Auxiliary Factor
This gene belongs to the splicing factor SR family of genes. U2 auxiliary factor, comprising a large and a small subunit, is a non-snRNP protein required for the binding of U2 snRNP to the pre-mRNA branch site. This gene encodes the small subunit which plays a critical role in both constitutive and enhancer-dependent RNA splicing by directly mediating interactions between the large subunit and proteins bound to the enhancers. Alternatively spliced transcript variants encoding different isoforms have been identified. [provided by RefSeq, Jul 2008]
USAF1, with 3.9 % occurrence is the 13th most common MPN mutation.